Introduction
Rhesus macaque, as a non-human primate model animal widely distributed in China, poses a unique model to explore the law of human evolution with the advantages of controllable environmental factors, convenient sampling and detection, and similar genome, physiological and pathological characteristics to humans. Because key issues regarding this animal model remain poorly addressed -- the limited genomic annotation and gene structure, lack of functional genomics data, and immature information research platform, rhesus macaque only has been mainly used in preclinical drug research for a long time, but not serve as a high-precision molecular medical model to support in-depth functional and mechanism studies.
We defined the rhesus macaque reference genome and gene structure in detail, established a rhesus macaque population genetics research platform, supplemented a large number of convincing transcripts missed before (72% of rhesus macaque transcripts were missed by existing authoritative annotations), and cleared the errors found in about 30% rhesus gene structure annotations from genomics platforms Ensembl and others. The self-produced and public data deeply integrated accounts for nearly 30% of the international non-human primate omics data. The breadth and depth of annotations for some regulatory levels are comparable to that of human genome annotations, which greatly enriches the understanding of rhesus macaque functional modulation.
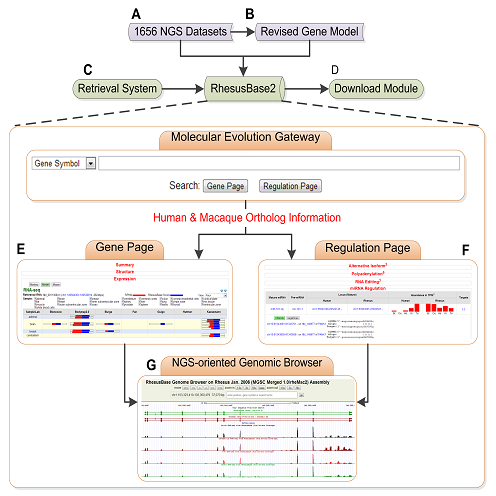
By further integrating 65 public databases and developing dozens of online interfaces, we built a one-stop genomic information knowledgebase, RhesusBase, which integrates annotation information of genome, gene structure, population genetics, expression regulation, molecular evolution, drug development, etc., totally with 7.6 billion functional annotation records. Especially, we developed Molecular Evolution Gateway, an NGS-oriented genomic framework to access and visualize macaque annotations in reference to human orthologous genes and the associated regulations, and PopGateway, the first genome-wide effort to identify and visualize the population genetics profile in rhesus monkey. Overall, RhesusBase provides a basis for precise functional and mechanism studies using rhesus macaque as a characteristic model.
RhesusBase News
More- [2023-01-02] (New) Nature Ecology & Evolution: De novo genes with an lncRNA origin encode unique human brain developmental functionality
- [2019-02-04] Genome Biology: Evolutionarily significant A-to-I RNA editing events originated through G-to-A mutations in primates
- [2017-10-12] (cover story) Molecular Biology and Evolution: Isoform Evolution in Primates through Independent Combination of Alternative RNA Processing
- [2016-03-21] RhesusBase PopGateway was featured by Molecular Biology and Evolution
- [2016-03-01] Molecular Biology and Evolution: The RhesusBase team release the population genetcis atlas in Rhesus Macaque
- [2015-08-31] The RhesusBase team reports a new mechanism for the origination of functional human-specific proteins
- [2015-06-28] Population Genetics Gateway released!
Community News
More- [2016-10-28] Nature: A comprehensive transcriptional map of primate brain development
- [2016-10-24] Nature: Mutations in Human Accelerated Regions Disrupt Cognition and Social Behavior
- [2016-02-25] Science: Single Antibody Protects Rhesus Macaques from Ebola
- [2016-01-26] Nature: Autism-like behaviours and germline transmission in transgenic monkeys overexpressing MeCP2
Explore RhesusBase
|

|
|
|
|

|

|
|
| STellaris | Gene Page | UCSC-mode Browser | Molecular Evolution | RhesusBase Quality Score | Drug Discovery | Frequently-used Files |
Citing RhesusBase
-
For Gene Page, Genome Browser, Molecular Evolution Gateway and Population Genetics Gateway:
Xiaoming Zhong et al. RhesusBase PopGateway: Genome-wide Population Genetics Atlas in Rhesus Macaque. Molecular Biology and Evolution, 2016. [PubMed]
Shi-Jian Zhang et al. Evolutionary Interrogation of Human Biology in Well-annotated Genomic Framework of Rhesus Macaque. Molecular Biology and Evolution, 2014. [PubMed]
Shi-Jian Zhang et al. RhesusBase: a Knowledgebase for the Monkey Research Community. Nucleic Acids Research, 2013. [PubMed]
-
For In-House NGS Data Preparation:
Xin-Zhuang Yang et al. Selectively Constrained RNA Editing Regulation Crosstalks with piRNA Biogenesis in Primates. Molecular Biology and Evolution, 2015. [PubMed]
Jia-Yu Chen et al. Emergence, Retention and Selection: A Trilogy of Origination for Functional De Novo Proteins from Ancestral LncRNAs in Primates. PLOS Genetics, 2015. [PubMed]
Jia-Yu Chen et al. RNA Editome in Rhesus Macaque Shaped by Purifying Selection. PLOS Genetics, 2014. [PubMed]
Chen Xie et al. Hominoid-specific De Novo Protein-coding Genes Originating from Long Non-coding RNAs. PLoS Genetics, 2012. [PubMed]