Mutations in Human Accelerated Regions Disrupt Cognition and Social Behavior
2016-10-24
Graphical Abstract
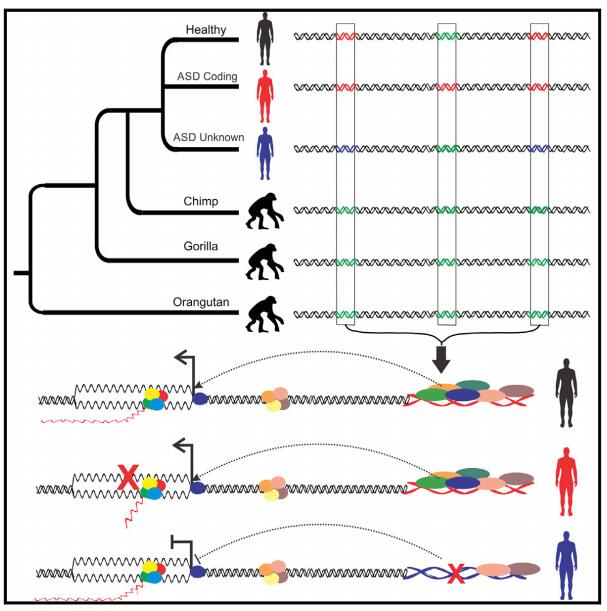
Small regions of the genome where humans have diverged from chimpanzees contain a variety of mutations implicated in autism and other neurodevelopmental disorders, report Harvard Medical School researchers at Boston Children's Hospital.
Published online Sept. 22 in Cell, the genomic analysis-the most comprehensive of its kind to date-opens a new approach to understanding both cognitive/behavioral disorders and the still-mysterious genetic changes that made human language, culture and civilization possible.
"This work brings together the study of evolution and the study of neurological disease," said Walsh,HMS Bullard Professor of Pediatrics and Neurology at Boston Children's and senior author of the Cell paper "Studying the kinds of mutations in HARs that cause neurodevelopmental disorders like autism spectrum disorder may tell us about the sorts of changes that led to us having a different brain than other animals'. Chimps are social creatures, but they're different from humans. They don't live in compact cities of a million people. That requires extraordinary social behavior."
For details, refer to the paper:
Mutations in Human Accelerated Regions Disrupt Cognition and Social Behavior. Cell,Volume 167, Issue 2, 6 October 2016, Pages 341-354.e12.Ryan N. Doan et al.
Abstract:
Comparative analyses have identified genomic regions potentially involved in human evolution but do not directly assess function. Human accelerated regions (HARs) represent conserved genomic loci with elevated divergence in humans. If some HARs regulate human-specific social and behavioral traits, then mutations would likely impact cognitive and social disorders. Strikingly, rare biallelic point mutations-identified by whole-genome and targeted "HAR-ome" sequencing-showed a significant excess in individuals with ASD whose parents share common ancestry compared to familial controls, suggesting a contribution in 5% of consanguineous ASD cases. Using chromatin interaction sequencing, massively parallel reporter assays (MPRA), and transgenic mice, we identified disease-linked, biallelic HAR mutations in active enhancers for CUX1, PTBP2, GPC4, CDKL5, and other genes implicated in neural function, ASD, or both. Our data provide genetic evidence that specific HARs are essential for normal development, consistent with suggestions that their evolutionary changes may have altered social and/or cognitive behavior.