Get the sequence of gene
Question:
How can I get the sequence and the associated annotations of ENSMMUG00000016081?
Response:
With the new RhesusBase genome browser, users could easily access the sequences and functional annotations of interested genes according to the style of UCSC genome browser. Taking the gene ("ENSMMUG00000016081") as an example; the sequence and the associated annotations could be found when following the steps as follows:
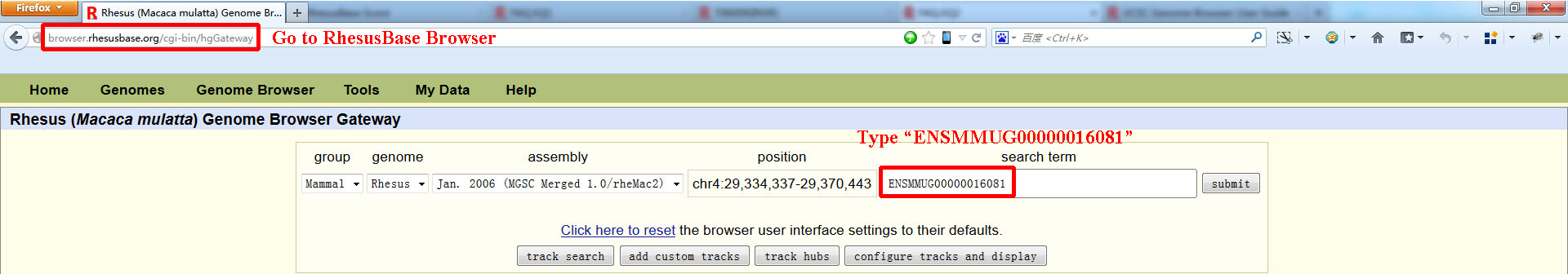
- Go to RhesusBase Genome Browser and type 'ENSMMUG00000016081' in the searching box as the input.
- When submit the query, the user will be linked to the genome browser showing the definite genomic region of this gene, in which more than eight functional categories (70 functional tracks) associated with the genomic regions of this gene were shown. In this case, 8 functional tracks were shown. The users could load other functional tracks following the style of UCSC genome browser.
- To get the sequences of this gene (“ENSMMUG00000016081”), the users could click the entry in 'Ensembl Gene Predictions – Ensembl 68' Track to enter the specific information page for this entry.
- Then the protein sequence, predicted mRNA sequence, as well as the genomic sequence near this gene are available in FASTA format in the section of 'Links to sequence'.
- The users could also get detailed annotations of this gene through the gene page. Briefly, from the homepage of RhesusBase, type 'ENSMMUG00000016081' in the searching box as the input.
- When submit the query, the user will be linked to the Gene Page of this gene, with detailed functional annotations in different categories, such as gene information, expression profile, regulation, variation and repeats, phenotypes and disease, function, as well as drug development.