Use RhesusBase Quality Score
Question:
How to use the RhesusBase Quality Score?
Response:
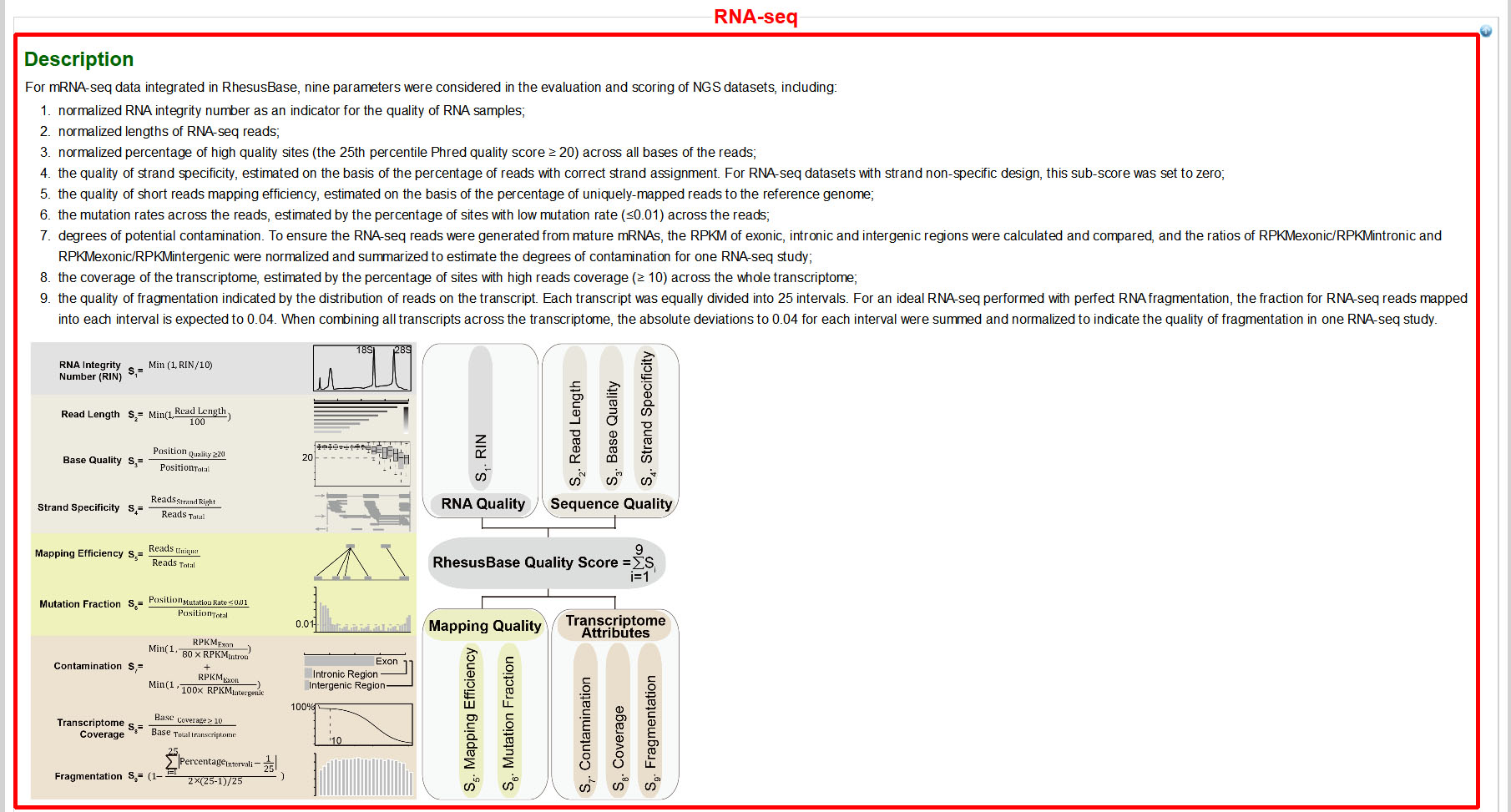
To facilitate NGS data assessment and comparison, a RhesusBase quality score (ranging from 0-10) was assigned to each NGS dataset according to multiple parameters that consider overall workflow of data generation and the downstream computational processing. Considering the differences of the sequencing platforms, experimental designs and data qualities, we also provided the percentile rank of the RhesusBase quality score among the same category of NGS dataset, defined as the percentage of scores in its frequency distribution that are the same or lower than it in all NGS datasets of this type, to facilitate RhesusBase users to set their own thresholds for NGS data quality control (ranging from 0-100). Generally NGS datasets with higher RhesusBase quality score or higher percentile rank are more reliable. Here is an example for the interpretation of RhesusBase quality score:
- For a specific RNA-Seq dataset (“Kaessmann_male2_testis”), the RhesusBase quality score and the percentile rank of the RhesusBase quality score are shown in Gene Pages with the RPKM scores of the gene. In this case, the RhesusBase quality score for this NGS dataset (“Kaessmann_male2_testis”) is 7.29, a quality score higher than 81.96% of RNA-seq datasets archived in the current version of RhesusBase.
- More detailed information for the generation and assignment of RhesusBase quality scores to NGS datasets is available on RhesusBase website:
- In this page, parameters (ranging from two to nine parameters for different types of NGS dataset) used to evaluate each type of NGS dataset were summarized in detail.
- For each NGS dataset, sub-scores for each evaluation were also shown with descriptions. In this case (“Kaessmann_male2_testis”), although both the the RhesusBase quality score and the percentile rank are high, the sub-score for the evaluation of “Strand Specificity” is low. This strand nonspecific RNA-seq dataset should thus be used with caution when analyzing overlapping genes (such as cis-natural antisense transcript).